FAQ
1. When is CMBD used?
The CMBD integrates comprehensive information of metabolic biomarkers for human cancers. It will be a useful tool in these cases: 1) retrieving metabolic biomarkers for designated sites, usage, class and flag; 2) searching specific biomarkers and its detail information reported in the literatures; 3) researching metabolic phenotypes of cancers; 4) seeking the mechanisms and management of cancers with systems biology approach
2. How is data standardized and annotated?
The data entry is shown in Unified Modeling Language (UML) class diagram (Figure 1). The cancer’s specific names and International Classification of Disease (ICD, 10th revision) codes are included to clarify disease information. Combined biomarkers are recorded in custom forms to help remember and understand them easily, such as AUC-CSH, TS×DPD, model_28389631_4_components. Their components are also recorded for further study. Single molecular biomarkers’ names and public entry ID are included to avoid duplicate records because of synonyms. Their detail information is quoted from corresponding public database or literature. Examples of single molecular biomarkers standardization and annotation are shown in table1
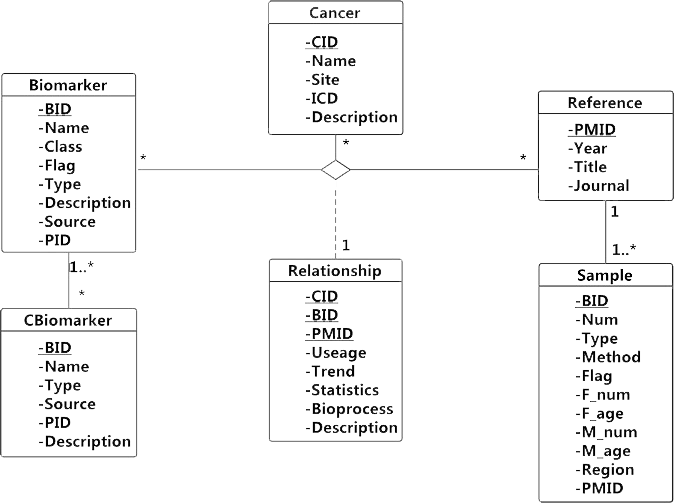
Figure 1:UML class diagram
Table 1 Examples of biomarkers standardization and annotation

3. How to use CMBD?
CMBD boasts a user-friendly web interface (Figure 2). In the ‘browse’ page, users have the option to refine their search parameters using the filters (site, class, usage and flag) as needed and then click the ‘Browse’ button (Figure 2a); items matching the filter criteria will be listed in a new page and users can access detailed biomarker information by clicking the ‘More’ link (Figure 2b). General information is contained on this page, in addition to detailed information in relation to cancer, biomarkers, sample type and references; clicking the link brings the user to the corresponding public database (Figure 2e). In the ‘search’ page, users can query the database using the biomarker name (Figure 2c). Results for fuzzy retrieval are also listed in a new page and users can get additional information in relation to these hits by click the ‘More’ link (Figure 2d). Users can download or submit data in the ‘Download’ or ‘Submit’ page, respectively.
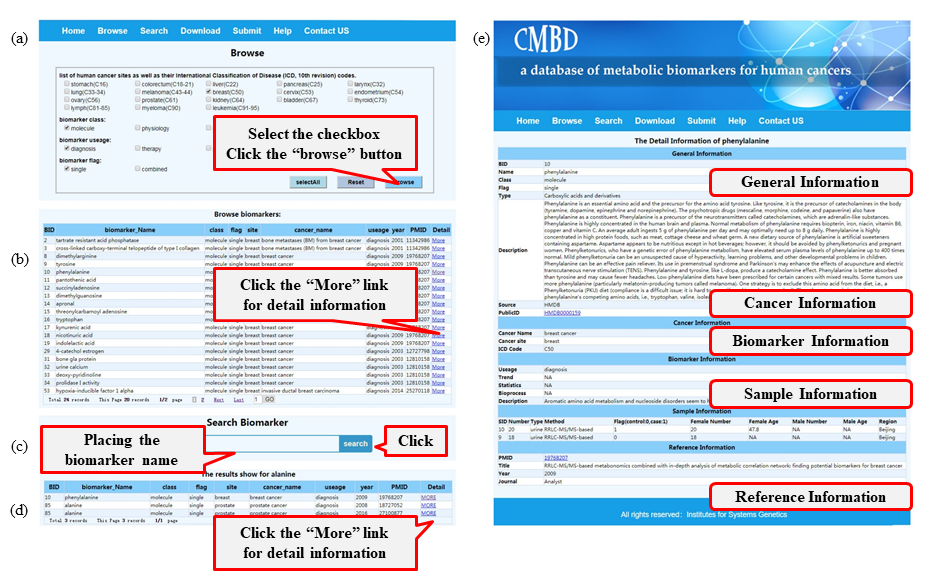
Figure 2:A screenshot of the utility of CMBD